DINEOF
好多人在遥感数据重构中会用到DINEOF方法,但是很多人不会用,这里记录笔记和教程。
官网: http://modb.oce.ulg.ac.be/mediawiki/index.php/DINEOF
下载
从网站下载压缩包解压之后,有如图的目录结构,一般只需要用到框出来的这几样东西。
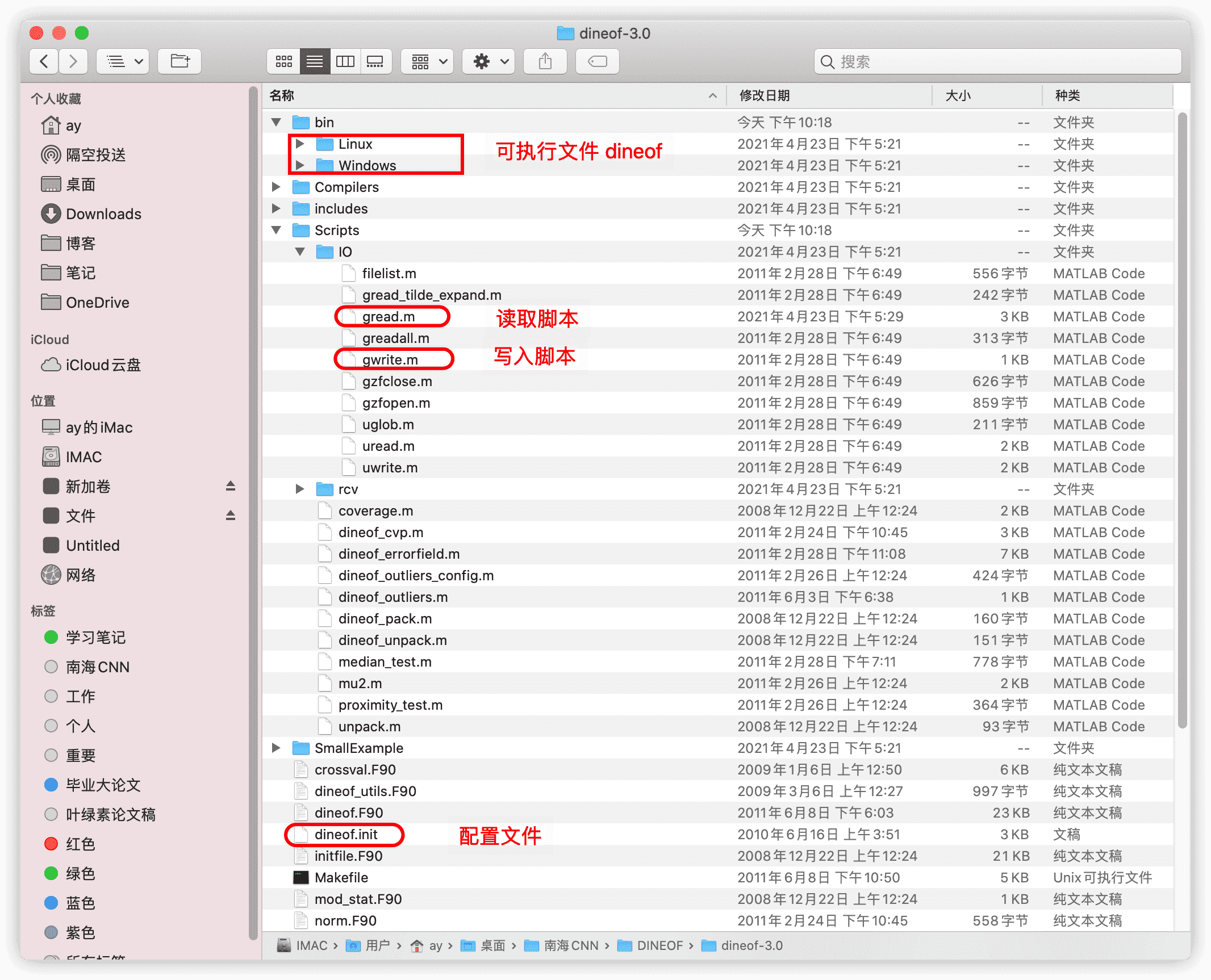
将下载文件夹里需要的文件复制到工作目录下,然后进行以下操作。
读取数据
基本步骤:
- 读取自己的文件,整理三个东西,分别是你要插值的变量,陆地掩码值,日期;
- 利用下载的dineof中的Matlab函数gwrite写成dineof可以直接读的数据
.dat格式 - 写好配置文件
dineof.init用shell脚本运行dineof dineof.init
%% 南海叶绿素 元数据 写成DINEOF的格式
filename = 'CCI_ALL-v5.0-DAILY.nc';
ncinfos= ncinfo(filename);
chlor_a = ncread(filename,'chlor_a');
lon = ncread(filename,'lon');
lat = ncread(filename,'lat');
time = ncread(filename,'time');
% generate mask
[m,n,k] = size(chlor_a)
mask = ones(m,n);
for i = 1:m
for j = 1:n
if sum(isnan(chlor_a(i,j,:)))>k-15
mask(i,j) = 0;
end
end
end
gwrite('./occi14dat/chla.dat',chlor_a);
gwrite('./occi14dat/mask.dat',mask);
gwrite('./occi14dat/date.dat',time);
配置文件
有几个点解释一下:
- 陆地掩码:0-1矩阵,将数据中
NaN,或者缺失率超过一定比例的就设置为0,需要插值的海域设置为0. - 时间序列:生成一个代表时间连续性的时间列,不需要日期,只需要表示时间是否连续即可。比如说是一个月的天数据,那么时间就是1-30即可。如果中间缺失某一天,那么连续值需要跳过。
- 配置文件
dineof.init:# ! 表示注释,不执行,我们需要将matlab生成的dat文件路径在配置文件中详细写明确。分别为data=[]mask=[]time=[]还有输出放在哪个文件夹里output=[],除此之外,还有一些详细的参数,至于这些参数就需要读者理解原理,根据具体的任务和计算资源来进行调整了。
!
! INPUT File for dineof 2.0
!
! Lines starting with a ! or # are comments
!
! gappy data to fill by DINEOF. For several matrices, separate names with commas
! Example:
! data = ['seacoos2005.avhrr','seacoos2005.chl']
data = ['./occi14dat/chla.dat']
!data = ['2Dbelcolour_region_period_anomaly.gher']
!data = ['2Dbelcolour_region_period_dat.gher']
! land-sea mask of gappy data. Several masks separated by commas:
! Example :
! mask = ['seacoos2005.avhrr.mask','seacoos2005.chl.mask']
mask = ['./occi14dat/mask.dat']
!mask = ['transpmaskindineof1D.gher']
time = './occi14dat/date.dat'
alpha = 0.01
numit = 3
!
! Sets the numerical variables for the computation of the required
! singular values and associated modes.
!
! Give 'nev' the maximum number of modes you want to compute
nev = 5
! Give 'neini' the minimum number of modes you want to compute
neini = 1
! Give 'ncv' the maximal size for the Krylov subspace
! (Do not change it as soon as ncv > nev+5)
! ncv must also be smaller than the temporal size of your matrix
ncv = 10
! Give 'tol' the treshold for Lanczos convergence
! By default 1.e-8 is quite reasonable
tol = 1.0e-8
! Parameter 'nitemax' defining the maximum number of iteration allowed for the stabilisation of eofs obtained by the cycle ((eof decomposition <-> truncated reconstruction and replacement of missing data)). An automatic criteria is defined by the following parameter 'itstop' to go faster
nitemax = 300
! Parameter 'toliter' is a precision criteria defining the threshold of automatic stopping of dineof iterations, once the ratio (rms of successive missing data reconstruction)/stdv(existing data) becomes lower than 'toliter'.
toliter = 1.0e-3
! Parameter 'rec' for complete reconstruction of the matrix
! rec=1 will reconstruct all points; rec=0 only missing points
rec = 1
! Parameter 'eof' for writing the left and right modes of the
!input matrix. Disabled by default. To activate, set to 1
eof = 1
! Parameter 'norm' to activate the normalisation of the input matrix
!for multivariate case. Disabled by default. To activate, set to 1
norm = 0
! Output folder. Left and Right EOFs will be written here
!
!Output = './'
Output = 'Output14/'
!
! user chosen cross-validation points,
! remove or comment-out the following entry if the cross-validation points are to be chosen
! internally
!
! clouds = 'crossvalidation.clouds'
!
! "results" contains the filenames of the filled data
!
!results = ['All_95_1of2.sst.filled']
!results = ['Output/F2Dbelcolour_region_period_datfilled.gher']
results = ['./Output14/chla_filled.nc']
! seed to initialize the random number generator
seed = 243435
!-------------------------!
! cross-validation points !
!-------------------------!
!number_cv_points = 7000
!cloud surface size in pixels
cloud_size = 500
!cloud_mask = 'crossvalidation.mask'
!
! END OF PARAMETER FILE
!
运行DINEOF
配置好dineof.init文件之后,就可以在shell中通过如下命令来跑模型:
cd ./
./dineof dineof.init
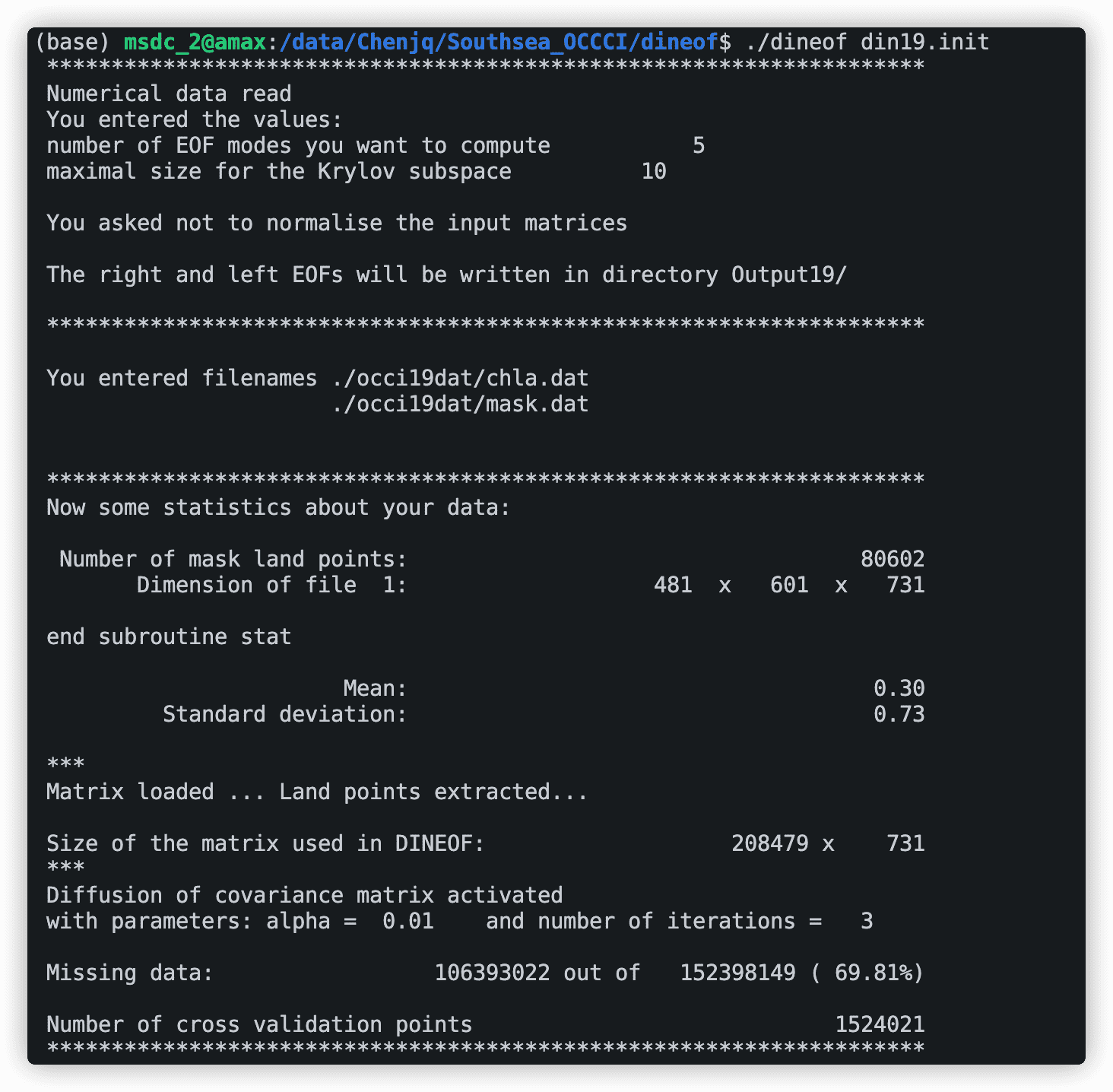
刚开始会输出一些你的输入信息,包括输入的文件和数据大小,同时还会计算数据的缺失率,后面就是模型的迭代过程了。
运行结束后,输出文件保存在Output/文件夹下,可以用Matlab通过下载的脚本里面的gread来调用,查看dineof插值之后的数据。
%% 读取DINEOF之后的数据
chla_filled = gread('Output/chla_filled.nc');
下载地址
好像官方不开放了,我看最近需要这个的人比较多,给大家分享云盘链接哈,大家帮忙点个赞哈哈 (只是为了填充数据库哈哈,没有流量也不会变现)
dineof-3.0
https://www.aliyundrive.com/s/CjKom7qdpPq
点击链接保存,或者复制本段内容,打开「阿里云盘」APP ,无需下载极速在线查看,视频原画倍速播放。

